 | |||||
|
| |||||
|
WELCOME TO THE ESSAY ARCHIVES!
<< June
<< Part I
(June/July)
<< June
<< June
<< June
<< Part I
(June/July)
* * *
|
Where Scrabble Meets Lego in the Building Blocks of Life: Synthetic Biology Researchers at the J. Craig Venter Institute (JCVI), a not-for-profit genomic research organization, published
results today describing the successful construction of the first self-replicating, synthetic bacterial cell . . . The synthetic cell is called Mycoplasma mycoides JCVI-syn 1.0 and is the proof of principle that genomes can be designed in the computer,
chemically made in the laboratory and transplanted into a recipient cell to produce a new self-replicating cell controlled only by the synthetic genome." JCVI press release, May 20, 2010 (n1) [It's] "the first self-replicating species we've had on the planet whose parent is a computer." Craig Venter of JCVI at press conference describing the JCVI cell (n2) Imagine, for a second, that you are given two boxes. In one are scrabble letters, but only the letters A, T, G and C. In the other are Lego pieces of widely varying shapes and sizes. Your task is to build something so specific that it can only work if you get the exact combination of letters and pieces correct. You're allowed to use a computer, however, since the words might be so long and the pieces so complex that only a computer could design them correctly. If you get everything just right, your creation will come to life and do exactly what you want it to. If you can picture this, they you are on the road to understanding the new and growing field of synthetic biology. Last year's announcement by scientists at the J. Craig Venter Institute (JCVI) that they had created the first self-replicating synthetic bacterial cell prompted both an increasing awareness of the field of synthetic biology and a call from President Barack Obama for a study of the implications of the scientific milestone. The study, conducted by the Presidential Commission for the Study of Bioethical Issues and entitled "New Directions: The Ethics of Synthetic Biology and Emerging Technologies," was released in December 2010 and is available at www.bioethics.gov. Some experts in the field say that the JCVI achievement was not a "game changer" because the scientists "had not actually created a synthetic life form . . . [but] had borrowed a naturally occurring cell and inserted a synthetic version of a naturally-occurring genome." (n3) However, others say that it is a "game-changing" accomplishment because "the main point of making synthetic cells is to make new kinds of cells - cells that perform useful and desired functions beyond the capability of any existing form of life . . . [and] the JCVI achievement opens the door to this possibility." (n4) This month's essay will take a brief look at the field of synthetic biology, provide background to aid in understanding what the JCVI scientists accomplished, review the conclusions of the Commission's report, and look briefly at the directions in which future research in the field of synthetic biology might lead. What is Synthetic Biology? Because of the early stage of development of the field and its interdisciplinary nature, there is not one single definition of the field of synthetic biology. However, for the purposes of the Presidential Commission's study, it was defined as follows: Synthetic biology is the name given to an emerging field of research that combines elements of biology, engineering, genetics, chemistry, and computer science. The diverse but related endeavors that fall under its umbrella rely on chemically-synthesized DNA, along with standardized and automatable processes, to create new biochemical systems or organisms with novel or enhanced characteristics. Whereas standard biology treats the structure and chemistry of living things as natural phenomena to be understood and explained, synthetic biology treats biochemical processes, molecules, and structures as raw materials and tools to be used in novel and potentially useful ways, often quite independent of their natural roles. It joins the knowledge and techniques of biology with the practical principles and techniques of engineering. "Bottom-up" synthetic biologists, those in the very earliest stages of research, seek to create novel biochemical systems and organisms from scratch, using nothing but chemical reagents. "Top-down" synthetic biologists, who have been working for several decades, treat existing organisms, genes, enzymes and other biological materials as parts or tools to be reconfigured for purposes chosen by the investigator. (n5) In its essence, synthetic biology "seeks to enable a new paradigm of biology by design" (n6) in which a desired biological function is conceived, designed and engineered, and then built to work in a specific way. (n7) Central to the process are "computational tools and rigorous quantitative methods to help design [the] configuration that will perform [the desired function] . . . [moving us] from what does exist to what can exist." (n8) Further information on the field of synthetic biology, including a "Synthetic Biology 101" introduction, links to online resources, and an index of selected readings in the field can be found at www.synbioproject.org. If you'd like to test yourself on a quick quiz on synthetic biology before continuing, click here. DNA, The Genome and the Cell: A Brief Review The synthetic genome created at JCVI is nearly the same as that of the genome of the natural bacterium, but the feat of its creation was neither a simple nor inexpensive endeavor. The accomplishment came at a cost of about $40 million with a team more than 20 scientists working on it for nearly 15 years. (n9), (n10) To more fully understand exactly what the JCVI scientists did, and how their work differs from the techniques of biotechnology, a brief review of DNA, the genome and the cell will be presented. (If necessary, a link to a glossary of related terms can be reached by clicking here for English and clicking here for Spanish.)
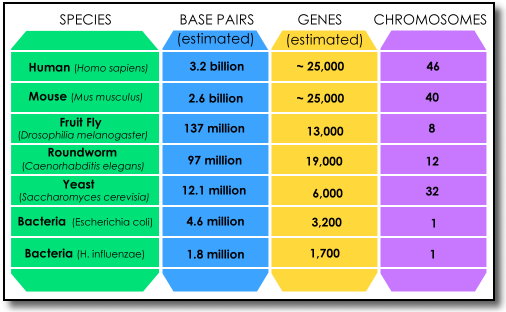
From the genome to the proteome. Photo credit: U.S. Department of Energy, Genomic Sciences Program, Biological and Enviromental Research Information System (BERIS), http://genomicscience.energy.gov or http://science.energy.gov/ber. As illustrated in the picture above, cells are the basic working units of every living system. All the instructions needed to direct their activities are contained within the chemical DNA (deoxyribonucleic acid). DNA from all organisms is made up of the same physical and chemical components, known as bases. The four bases are Adenine (A), Thymine (T), Guanine (G) and Cytosine (C). These bases "pair" with each other in DNA; "A" always pairs with "T" and "G" always pairs with "C" in two chains known as a double helix. The DNA sequence is the particular side-by-side arrangement of bases along the DNA strand. This order spells out the exact instructions required to create a particular organism with its own unique traits. The genome is an organism's complete set of DNA, and genomes vary widely in size. The smallest known genome for a free-living organism (a bacterium) contains about 600,000 DNA base pairs. Human and mouse genomes have roughly three billion base pairs. All human cells except for red blood cells contain a complete genome. DNA in the human genome is arranged into 24 distinct chromosomes -- physically separate molecules that range in length from about 50 million to 250 million base pairs. Each chromosome contains many genes, which are the basic physical and functional units of heredity. Genes are specific sequences of bases that encode instructions on how to make proteins. The human genome is estimated to contain about 25,000 genes. Genes comprise only about two percent of the human genome; the remainder consists of noncoding regions, whose functions may include providing chromosomal structural integrity and regulating where, when, and in what quantity proteins are made. Proteins perform most life functions and even make up the majority of cellular structures. Proteins are large, complex molecules made up of smaller subunits called amino acids. Chemical properties that distinguish the 20 different amino acids cause the protein chains to fold up into specific three-dimensional structures that define their particular functions in the cell. The sum total of all proteins in a cell is know as the proteome. (n11), (n12) Today's ability to construct or reconstruct the genome of a cell owes much to the knowledge and insight gained through the Human Genome Project. The Human Genome Project was a 13-year effort which began in 1990. Goals of the Human Genome Project (www.ornl.gov/sci/techresources/Human_Genome/home.shtml) were to: identify all of the approximately 20,000 - 25,000 genes in a human body, determine the sequences (or order) of the three billion chemical base pairs that make up human DNA, store the information in databases, improve tools for data analysis, transfer related technologies to the private sector and address the ethical, legal and social issues arising from the project. (n13) In addition to sequencing the complete human genetic code, researchers also sequenced the genomes of non-human organisms like bacteria, fruit flies and laboratory mice. The chart below shows the differences in numbers of chromosomes, genes and base pairs in a number of species whose genomes have been sequenced.
Chart derived from information on U.S. Department of Energy, Human Genome Project Information site, Functional and Comparative Genomics Fact Sheet. Viewed online August 2011 at www.ornl.gov/sci/techresources/Human_Genome/project/faq/compgen.shtml Human Genome Project research within the U.S. Department of Energy was directed by Aristides Patrinos, head of the Office of Biological and Environmental Research, and Human Genome Project activities with in the National Institutes of Health (NIH) were directed by Francis Collins. Craig Venter was originally part of this Human Genome Project effort but left to direct parallel research activities as head of the private company Celera Genomics and went on to found the J. Craig Venter Institute (JCVI). In general, the ability to sequence DNA was followed by the development of techniques to do the reverse - to chemically construct - or synthesize - DNA from individual chemicals. According to the Bioethics Commission's report, "within the last few years, researchers have developed methods to accurately synthesize increasingly longer segments of DNA and to bring them together in even larger segments of DNA . . . Costs for DNA synthesis have fallen dramatically, . . . [and] computer modeling, not readily available until recently, [has facilitated] the design of novel genetically engineered biological systems . . . Although biological systems are not as easily modeled as an electronic circuit or a bridge, at least at this time, sophisticated simulations, mostly in single-celled systems [such as bacteria] are contributing to improved modeling of synthetic biological systems." (n14) It is this type of computer modeling which allowed the JCVI researchers to use digital information to design the synthetic genome which replaced the DNA in the bacterial cell - which perhaps makes the comment about the self-replicating species "whose parent is a computer" a bit easier to understand. The First Self-Replicating Bacterial Cell In essence what the JVCI researchers accomplished was to transform one natural bacterial cell into a different self-replicating bacterial cell with a synthetic genome, or to "reprogram" an existing cell to form new cells specified by the synthetic DNA. (n15) This was accomplished in several steps. First scientists began with an accurate, digitized genome of the bacterium M. mycoides. (n16) The team then designed 1078 specific cassettes of DNA that were 1080 base pairs long (n17) (a cassette is as sequence of DNA encoding one or more genes for a single biotechnical function. The ends of each cassette [in this case] overlapped by 80 base pairs in order for them to "stick" together). (n18) The cassettes were assembled in three stages within a yeast cell in progressively larger segments until the complete genome had been synthesized. The synthetic M. mycoides genome was then isolated from the yeast cell and transplanted into the recipient cell of a closely-related bacterium, Mycoplasma capricolum. (n19) The Mycoplasma capricolum cells had been treated in a way to allow replication following the natural cell cycle, (n20) and within two days only M. mycoides cells containing the synthetic DNA were visible on the petri dishes containing the bacterial growth medium. (n21) In much the same way that a photographer might watermark his pictures to make them more identifiable or prevent them from being copied, the JCVI scientists had inserted specific strings of bases into the synthetic DNA sequences which had been constructed. "Four of the ordered sequences contained strings of bases that, in code, spell[ed] out an e-mail address, the names of many of the people involved in the project, and a few famous quotations." (n22) The watermarks allowed the DNA of the new cells to be identified as synthetic - and distinguishable from the cell's natural DNA - upon sequencing. (n23) Synthetic Biology vs. Biotechnology There are those who would question whether accomplishments such as the synthetic genome are the start of a "new industrial revolution" in synthetic biology, or merely "old wine in new bottles." (n24) Since the means of producing a number of truly viable products via synthetic biology doesn't exist at this time, others claim that many companies ". . . have already come a long way with old-fashioned biotechnology, . . . [with] a large potential for developing [products] with the current technology." (n25) Most people would probably ask "Is there a difference between biotechnology and synthetic biology?" The answer to that question is "Yes." The use of biotechnology involves the "repeated manipulation of single [or small numbers] of [existing] genes" (n26) for the genes to express a certain trait or produce a desired result. Many people are familiar with genetically engineered crops such as corn, soy or rice (See also the 2004 Essays in the Essay Archives section). The biotechnology used in developing many of these crops, for example, "involves adding or altering less than ten genes out of the tens of thousands that are contained in most organisms or plants. Synthetic genomics is different in that scientists start with digital information, which allows for the design of entire synthetic chromosomes to replace existing chromosomes in cells." (n27) In other words, while biotechnology is concerned primarily with the manipulation of existing genetic material, synthetic biology is concerned primarily with the engineering of new genetic material. One example of an area in which biotechnology is being used to produce viable products - and synthetic genomics is being considered for future ones - is in renewable fuels, more specifically renewable fuel production from algae.
Diagram source: U.S. Department of Energy, Office of Energy Efficiency and Renewable Energy, Biomass Program, National Algal Biofuels Technology Roadmap, Washington, D.C.: U.S. DOE, 2010, p. vi Venter is also founder and CEO of another company called Synthetic Genomics (SGI, www.syntheticgenomics.com). His company recently entered into an agreement with ExxonMobil for research and development of biofuels from algae. Researchers in the program will be evaluating "various algae, including both natural and engineered strains, in . . . different growth systems under a wide range of conditions." (n30) If the research and development milestones are successfully met, ExxonMobil says it expects to spend more than $600 million on the algal biofuels program over the next decade, of which $300 million will be allocated to SGI. (n31) To view a 2011 ExxonMobil brochure on the program, click here, or go to www.exxonmobil.com/corporate, then click on "Media," and "Publications," and scroll down to the pdf file for Algal Biofuels. To learn a bit more about how algal biodiesel works, visit http://science.howstuffworks.com/environmental/green-science/algae-biodiesel.htm. "Top-Down" and "Bottom-Up" Approaches to Synthetic Biology As mentioned earlier, the Presidential Commission's report identified the two main approaches to synthetic biology as being "top-down" and "bottom-up." In the top-down approach, "scientists use synthetic biology to redesign existing organisms or gene sequences with the goal of stripping out unnecessary parts, or replacing or adding specific parts to achieve new or amplified characteristics or functions. Using this approach, scientists aim to remove parts of an organism or genetic code to create what some have described as a 'chassis organism' that can then be modified through the addition or subtraction of engineered genetic circuits or metabolic pathways." (n32) The other more complex and challenging approach is the bottom-up approach which is as the name sounds - building "living systems from raw materials starting with non-living components." (n33) This approach is sometimes characterized by a reliance on "assembling systems from chemically-synthesized standard parts that perform desired functions," (n34) the set of existing parts most often compared to "Lego" pieces. The BioBricks Foundation (www.biobricks.org) houses an open catalog of these parts known as the Registry of Standard Biological Parts. The Registry is an "open catalog of standardized DNA parts that encode basic biological functions and can be easily combined and exchanged among different devices and laboratories. These standardized parts are made available to the public free of charge to further research in the field." (n35) There are links from the BioBricks Foundation website to the Parts Registry, synthetic biology conferences, labs and courses, and the 5th Meeting on Synthetic Biology which took place at Stanford in June of this year. A new generation of budding synthetic biologists also is becoming acquainted with this "bottom-up" approach through the annual iGEM, or International Genetically Engineered Machine, competition (www.igem.org). According to information on the iGEM website, the competition "is the premiere undergraduate synthetic biology competition," (n36) which was started in 2003. For the competition, student teams are given a kit of biological parts from the Registry of Standard Biological Parts at the start of the summer. Working at their own schools over the summer, they use these parts and parts of their own design to build biological systems and operate them in living cells [primarily bacteria]. (n37) iGEM contest projects in the past have included an RNA caffeine sensor, an arsenic bacteriosensor for drinking water, banana- and wintergreen- scented bacteria, synthetic multicellular bacteria, bactoblood (a system that produces hemoglobin in E coli and might some day be used as a replacement for red blood cells in emergency transfusions), and microbial biofuels. (n38) The competition has grown from 5 teams in 2004 to 165 teams in 2011 (n39), and there are links to the annual competition websites at www.igem.org. The Report of the Presidential Commission "Synthetic biology is thus a new topic in a growing debate about which . . . ideals should guide the human relationship to nature." (n40) "Genetic engineering can be described as softening unpleasant edges of the activity of an organism or plant, or as adding extra value to a plant or an organism beyond the value that it already has for us - beta-carotene enriched 'golden rice' as a case in point. In other words, if we look at nature through the glasses of genetic engineering, we see a world filled with entities that are already useful to us in many respects and that just need some reshaping here and there to perfectly match our interests. In contrast, synthetic biology does not soften edges, but creates life forms that are meant not to have any edges from the start. It does not add value to an existing organism; it brings into existence something that counts as valuable from our point of view. Seen from the perspective of synthetic biologists, nature is a blank space to be filled with whatever we wish." (n41) In a letter dated the same day as the JCVI researchers' announcement of their accomplishment, President Barack Obama requested that the Presidential Commission for the Study of Bioethics undertake "a study of the implications of this scientific milestone as well as other advances that may lie ahead in this field of research." (n42) The President requested that the Commission review potential benefits and risks, as well as considering appropriate ethical boundaries, and that the study be completed within six months. As stated earlier, that report was released in December 2010 and can be found on the Commission's site at www.bioethics.gov. In addition to a review of the field and its history, the panel spoke with and heard testimony from a wide variety of constituent groups and science and industry representatives, outlined areas in which synthetic biology might provide useful benefits (such as energy, biofuels, pharmaceuticals, health and biosecurity), and identified basic ethical principles for guiding the assessment of an emerging technology such as synthetic biology. What the Commission found, and stated in the executive summary of the report "is that the Venter Institute's research and synthetic biology are in the early stages of a long continuum of research in biology and genetics. The announcement [in] May, although extraordinary in many ways, does not amount to creating life as either a scientific or moral matter . . . The Commission therefore focused [in the report] on the measures needed to assure the public that these efforts [in synthetic biology] proceed with appropriate attention to social, environmental and ethical risks." (n43) The five principles identified by the Commission as being most relevant to assessing ethical considerations related to synthetic biology and other emerging technologies, also the principles used in framing their recommendations are:
In general, the Commission stated that it believes the "potential promise of synthetic biology is immense." (n44) However, it also stated that achievements in the field like that of the Venter Institute "raise a host of complex and often controversial issues. Breakthroughs can help humankind in many ways, but they invariably carry some risks. . . Proponents of synthetic biology cite its potential to reduce our reliance on fossil fuels and transform medical care and human health, among other possible benefits. Critics express concerns about 'playing God,' threatening biodiversity and the organization and natural history of the species, demeaning and disrespecting the meaning of life, and threatening longstanding concepts of nature. With these unprecedented opportunities and achievements comes an obligation to consider carefully both the promise and potential perils that they could realize." (n45) The full list of the Commission's recommendations can be found in the report, recommendations which the Commission says will "provide a publicly accountable basis for ensuring that the field of synthetic biology advances . . . with processes in place to identify, assess, monitor and mitigate risks on an ongoing basis as the field matures." (n46) Since the JCVI's announcement and subsequent release of the Commission's report, discussion of the field of synthetic biology and its greater implications has continued to grow. The Commission's report suggests that open dialogue should be ongoing as the field develops and matures. An example of the ongoing dialogue on synthetic biology is the current (July-August 2011) issue of a journal called the Hastings Center Report, and this month's essay will conclude with a quote drawn from one of the articles within that issue: Make no mistake. It is one thing to conduct a research program to unlock the remaining molecular secrets of life. It is quite another thing to program synthetic cells to do whatever we want. There is nothing stopping us from embarking on the research program. But today nobody has any idea how to program a partly or fully synthetic cell to do whatever we want - neither the JCVI team nor anybody else. If we could, then we already would have reprogrammed bacteria to produce inexpensive fuels, food, building materials, pharmaceuticals, you name it. People are actively working on these tasks, and no doubt many will eventually succeed. But today we can program only pretty trivial traits, and only one synthetic biology reprogramming project has had notable commercial success (making cheaper malarial drugs). Synthetic biology has reprogrammed bacteria to do many things, but progress is painstaking and each achievement requires surmounting many challenges. Furthermore, the difficulty increases by leaps and bounds as we try to reprogram much more complex traits. The JCVI team unlocked the door to arbitrarily reprogramming simple life forms, but figuring out how to go through that door and end up where we want remains a largely unsolved scientific challenge." (n47)
FOOTNOTES - The following are the footnotes indicated in the text in parentheses with the letter "n" and a number. If you click the asterisk at the end of the footnote, it will take you back to the paragraph where you left off. n1 - J. Craig Venter Institute press release, May 20, 2010, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/press-release-final.pdf (*) n2 - Wade, Nicholas, "Synthetic Bacterial Genome Takes Over a Cell, Researchers Report," New York Times, May 21, 2010, p. A17 (*) n3 - Kaebrick, Gregory E., "Synthetic Biology, Analytic Ethics," Hastings Center Report, Vol. 40, No. 4, July-August 2010, p. C3 (*) n4 - Bedau, Mark A., "The Intrinsic Scientific Value of Reprogramming Life," Hastings Center Report, Vol. 41, No. 4, July-August 2011, p. 29 (*) n5 - Presidential Commission for the Study of Bioethical Issues (PSCBI), New Directions: The Ethics of Synthetic Biology and Emerging Technologies, PCSBI: Washington, D.C., December 2010, p. 36 (*) n6 - Prepared statement Joshua N. Leonard in 21st Century Biology, hearing before the U.S. Congress, House of Representatives, Committee on Science and Technology, Subcommittee on Research and Science Education, Serial No. 111-103, 111th Congress, 2nd Session, June 29, 2010, US GPO: Washington D.C., 2010 (*) n9 - Pennisi, Elizabeth, "Synthetic Genome Brings New Life to Bacterium," Science, Vol. 328, Issue 5981, 21 May 2010, p. 958 (*) n10 - Templer, Laura, "Synthetic Biology: Injecting New Life into the Chemical Industry/How Did Venter Create a Synthetic Genome?" Chemical Week, December 13, 2010, p. 24 (*) n11 - U.S. Department of Energy, Human Genome Program, Genomics and its Impact on Science and Society, a 2008 Primer, and "From the Genome to the Proteome - Basic Science," viewed online August 2011 at www.ornl.gov/sci/techresources/Human_Genome/project/info.shtml and August 2011 at www.ornl.gov/sci/techresources/Human_Genome/publicat/primer2001/1.shtml (*) n12 - Campbell, Neil A. Biology, 3rd Edition, Redwood City, CA: Benjamin/Cummings Publishing Company, 1993, p. 86 (*) n13 - U.S. Department of Energy, Genomic Sciences Program, Human Genome Project information. Viewed online August 2011 at www.ornl.gov/sci/techresources/Human_Genome/project/about.shtml(*) n14 - Presidential Commission for the Study of Bioethical Issues (PSCBI), pp. 40 - 43 (*) n15 - J. Craig Venter Institute, "First Self-Replicating Synthetic Bacterial Cell," Frequently Asked Questions (FAQ), viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/faq.pdf (*) n16 - J. Craig Venter Institute press release, May 20, 2010, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/press-release-final.pdf (*) n18 - Templer, Laura, "Synthetic Biology: Injecting New Life Into the Chemical Industry," Chemical Week, Vol. 172, No. 30, December 13, 2010, p. 24 (*) n19 - J. Craig Venter Institute press release, May 20, 2010, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/press-release-final.pdf (*) n20 - Templer, Laura, "Synthetic Biology," p. 24 (*) n21 - J. Craig Venter Institute press release, May 20, 2010, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/press-release-final.pdf (*) n22 - Pennisi, Elizabeth, "Synthetic Genome Brings New Life to Bacterium," Science, Vol. 328, Issue 5981, 21 May 2010, p. 958 (*) n23 - Gibson, Daniel, et. al., "Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome," Science, Vol. 329, Issue 5987, 2 July 2010, p. 56 (*) n24 - Marris, Claire, Rose, Nikolas, and Zhang, Joy Y. BIOS Working Paper #4: The Transnational Governance of Synthetic Biology, London: London School of Economics/Center for the Study of Bioscience, Biomedicine, Biotechnology and Society (BIOS), May 2011, p. 6 (*) n25 - Templer, Laura, "Synthetic Biology," pp. 24 - 25 (*) n27 - J. Craig Venter Institute, "First Self-Replicating Synthetic Bacterial Cell," Frequently Asked Questions (FAQ), viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/faq.pdf, p. 1 (*) n28 - Templer, Laura, "Synthetic Biology," pp. 24 (*) n29 - Information from Solazyme video "The Future of Oil," viewed online August 2011 at www.solazyme.com/videos (*) n30 - Synthetic Genomics, "ExxonMobil and SGI Advance Algae Biofuels Program with New Greenhouse," press release dated July 14, 2010, viewed online August 2011 at www.syntheticgenomics.com/media/press/071410.html (*) n32 - Presidential Commission for the Study of Bioethical Issues (PSCBI), p. 43 (*) n36 - iGEM website information, viewed online August 2011 at www.igem.org (*) n38 - Carlson, Robert H. Biology is Technology, Cambridge, MA: Harvard University Press 2010, pp. 88 - 96 (*) n39 - iGEM website information, viewed online August 2011 at www.igem.org (*) n40 - Kaebnick, Gregory, "Of Microbes and Men," Hastings Center Report 41, no. 4, July-August 2011, p. 26 (*) n41 - Boldt, Joachim, and Muller, Oliver, "Newtons of the Leaves of Grass," Nature Biotechnology, Vol. 26, No. 4, April 2008, p. 388 (*) n42 - Letter of President Barack Obama in Presidential Commission for the Study of Bioethical Issues (PSCBI), p. vi (*) n43 - Presidential Commission for the Study of Bioethical Issues (PSCBI), p. 3 (*) n44 - Presidential Commission for the Study of Bioethical Issues (PSCBI), p. 50 (*) n45 - Presidential Commission for the Study of Bioethical Issues (PSCBI), p. 21 (*) n46 - Presidential Commission for the Study of Bioethical Issues (PSCBI), p. 17 (*) n47 - Bedau, Mark A. "The Intrinsic Scientific Value of Reprogramming Life," Hastings Center Report 41, No. 4, July-August 2011, p. 30 (*) LINKS INCLUDED IN ESSAY - The following are links included in the essay.
BIBLIOGRAPHY - The following is the Bibliography for the August 2011 essay. 21st Century Biology, Hearing before the U.S. Congress, House of Representatives, Committee on Science and Technology, Subcommittee on Research and Science Education, Serial No. 111-103, 111th Congress, 2nd Session, June 29, 2010, US GPO: Washington D.C., 2010 Bedau, Mark A., "The Intrinsic Scientific Value of Reprogramming Life," Hastings Center Report 41, No. 4, July-August 2011, pp. 29 - 31 BioBricks Foundation. SB 5.0: The Fifth International Meeting on Synthetic Biology, Program Booklet, viewed online August 2011 at www.sb5.biobricks.org Boldt, Joachim, and Muller, Oliver, "Newtons of the Leaves of Grass," Nature Biotechnology, Vol. 26, No. 4, April 2008, pp. 387 - 389 Campbell, Neil A. Biology, 3rd Edition. Redwood City, CA: Benjamin/Cummings Publishing Company, 1993 Carlson, Robert H. Biology is Technology. Cambridge, MA: Harvard University Press 2010 European Commission, New and Emerging Science and Technology (NEST) High-Level Expert Group. Synthetic Biology: Applying Engineering to Biology. Luxembourg: Office for Official Publications of the European Community, 2005 Gibson, Daniel G., et al, "Creation of a Bacterial Cell Controlled by a Chemically Synthesized Genome," Science, Vol. 329, Issue 5987, 2 July 2010, pp. 52 - 56 Gutmann, Amy, "The Ethics of Synthetic Biology: Guiding Principles for Emerging Technologies," Hastings Center Report 41, No. 4, July-August 2011, pp. 17 - 22 International Genetically Engineered Machine (iGEM) competition Home Page, viewed online August 2011 at www.igem.org J. Craig Venter Institute. Press Release, First Self-Replicating Synthetic Bacterial Cell, May 20, 2010, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/press-release-final.pdf J. Craig Venter Institute. Freqently Asked Question (FAQ), First Self-Replicating Synthetic Bacterial Cell, viewed online August 2011 at www.jcvi.org/cms/fileadmin/site/research/projects/first-self-replicating-bact-cell/faq.pdf Fukuyama, Francis. Our Posthuman Future: Consequences of the Biotechnology Revolution. New York: Farrar, Straus and Giroux, 2002 Irvine, James H. and Schwarzbach, Sandra, "The Top 20 (Plus 5) Technologies for the World Ahead," The Futurist, Vol. 45, No. 3, May-June 2011, pp. 16 - 24 Kaebrick, Gregory E., "Of Microbes and Men," Hastings Center Report 41, no. 4, July-August 2011, p. 25 - 28 Kaebrick, Gregory E., "Synthetic Biology, Analytic Ethics," Hastings Center Report 40, No. 4, July-August 2010, p. C3 Marris, Claire, Rose, Nikolas, and Zhang, Joy Y. BIOS Working Paper #4: The Transnational Governance of Synthetic Biology. London: London School of Economics/Center for the Study of Bioscience, Biomedicine, Biotechnology and Society (BIOS), May 2011 Murray, Thomas A., "Interests, Identities and Synthetic Biology," Hastings Center Report 41, No. 4, July-August 2011, pp. 31 - 36 Pennisi, Elizabeth, "Synthetic Genome Brings New Life to Bacterium," Science, Vol. 328, Issue 5981, 21 May 2010, pp. 958 - 959 Presidential Commission for the Study of Bioethical Issues (PSCBI), New Directions: The Ethics of Synthetic Biology and Emerging Technologies. Washington, D.C.: U.S. Government Printing Office, December 2010 Solazyme. Video, "The Future of Oil," viewed online August 2011 at www.solazyme.com/videos Synthetic Genomics. Press Release: "ExxonMobil and Synthetic Genomics, Inc. Advance Algae Biofuels Program with New Greenhouse," dated July 14, 2010, viewed online August 2011 at www.syntheticgenomics.com/media/press/071410.html Templer, Laura, "Synthetic Biology: Injecting New Life into the Chemical Industry," Chemical Week, December 13, 2010, pp. 24 - 25 U.S. Department of Energy, Office of Energy Efficiency and Renewable Energy, Biomass Program, National Algal Biofuels Technology Roadmap. Washington, D.C.: U.S. DOE, 2010 U.S. Department of Energy, Human Genome Program. Genomics and its Impact on Science and Society, a 2008 Primer, and "From the Genome to the Proteome - Basic Science," viewed online August 2011 at www.ornl.gov/sci/techresources/Human_Genome/project/info.shtml and August 2011 at www.ornl.gov/sci/techresources/Human_Genome/publicat/primer2001/1.shtml U.S. Department of Energy, Genomic Sciences Program, Human Genome Project information. Viewed online August 2011 at www.ornl.gov/sci/techresources/Human_Genome/project/about.shtml Wade, Nicholas, "Synthetic Bacterial Genome Takes Over a Cell, Researchers Report," New York Times, May 21, 2010, p. A17
To return to the top of the page, click here. To return to the essay archives, click here. Follow www.dorothyswebsite.org on TWITTER! Home | Essays |
Poetry | Free Concerts | Links | 2011 Extras |
About the Site |
||||
 | |||||
|
www.dorothyswebsite.org © 2003 - 2011 Dorothy A. Birsic. All rights reserved. Comments? Questions? Send an e-mail to: information@dorothyswebsite.org | |||||






 In Chess, as with many games with roots in more ancient times, the exact origin of the game is somewhat obscure. Historians
generally consider shatranj, developed in the Persian Empire around 600 A.D., to be one of the most direct ancestors of modern chess. Shatranj, however, grew to be a more refined version of
the earlier Indian games of chaturanga (a Sanskrit name for a battle formation) and ashtapada.
In Chess, as with many games with roots in more ancient times, the exact origin of the game is somewhat obscure. Historians
generally consider shatranj, developed in the Persian Empire around 600 A.D., to be one of the most direct ancestors of modern chess. Shatranj, however, grew to be a more refined version of
the earlier Indian games of chaturanga (a Sanskrit name for a battle formation) and ashtapada.